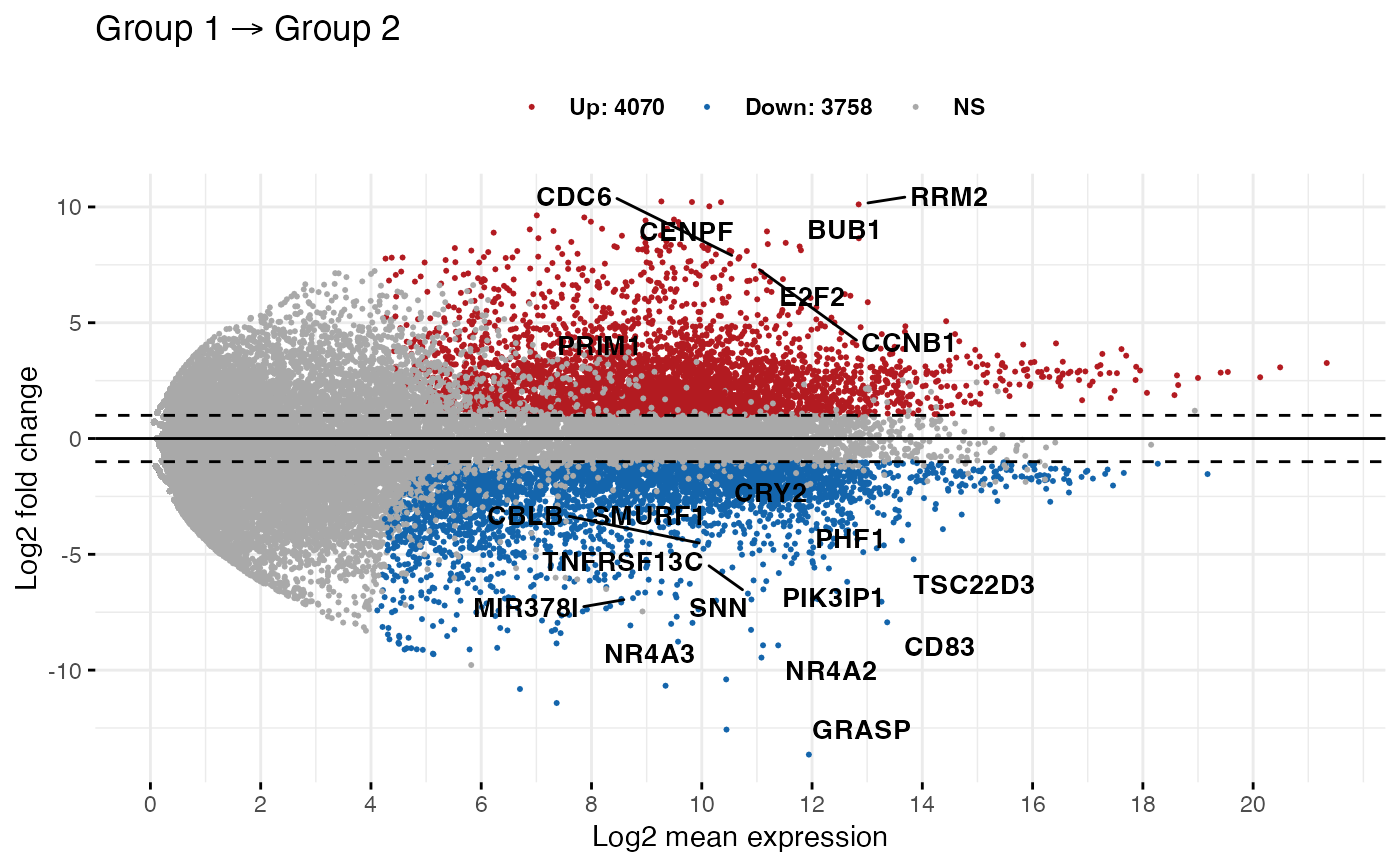
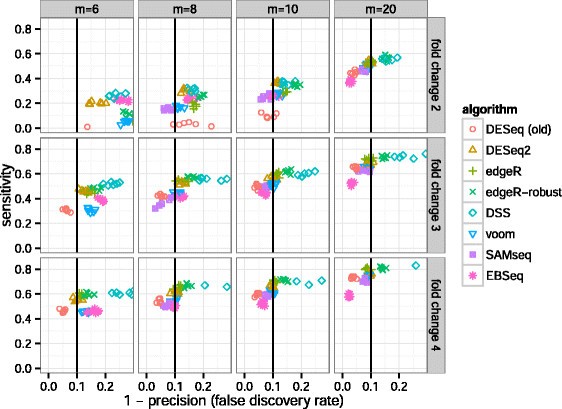
MD plot showing the log-fold change and average abundance of each gene.... | Download Scientific Diagram
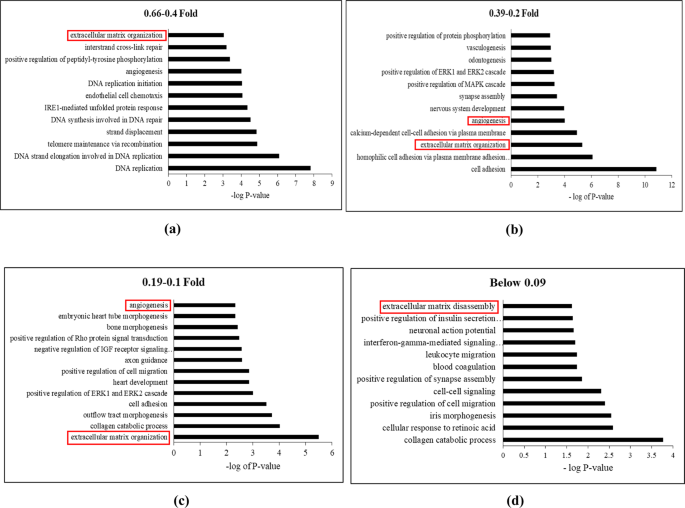
Transcriptomic Profiling of Adipose Derived Stem Cells Undergoing Osteogenesis by RNA-Seq | Scientific Reports
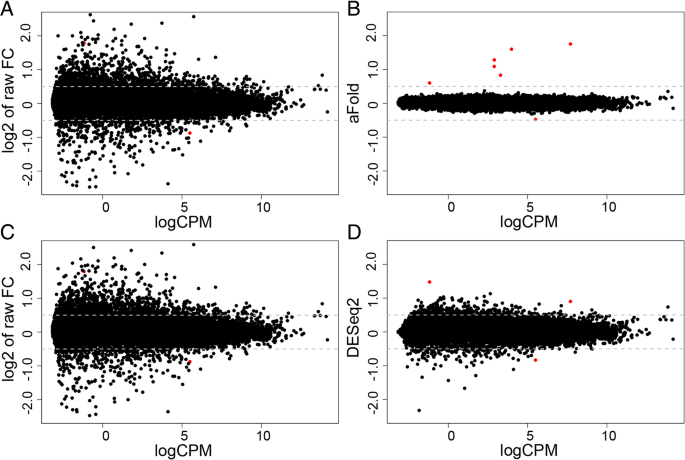
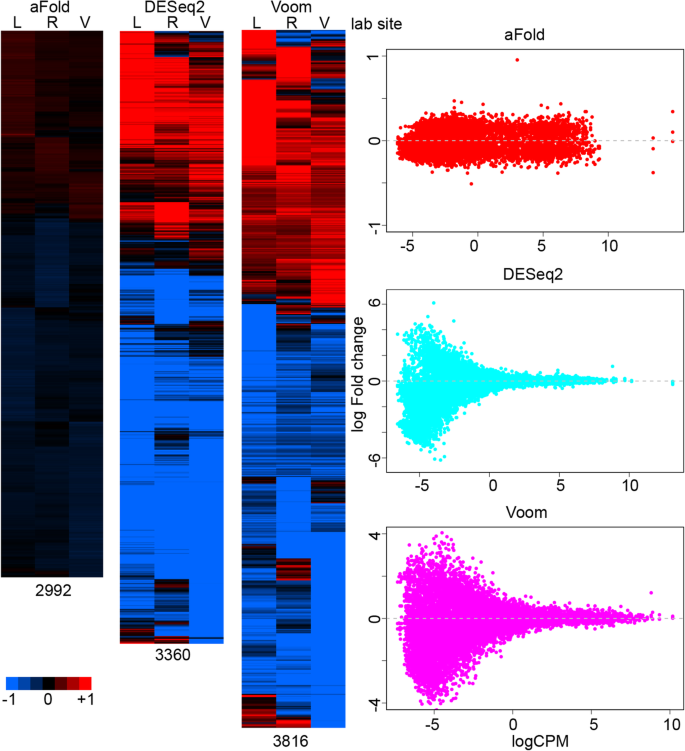
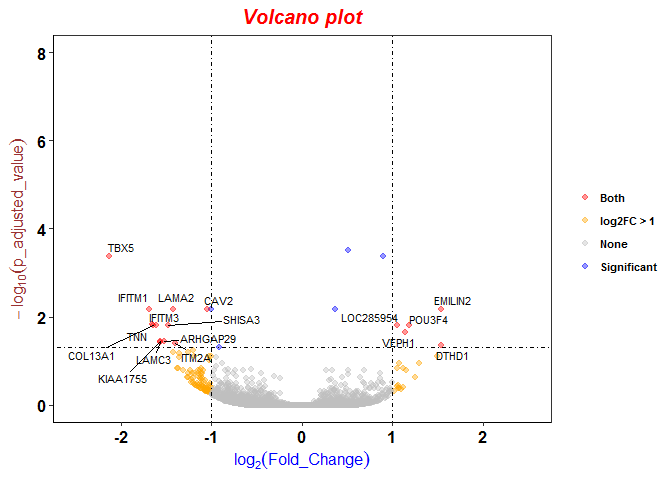
aFold – using polynomial uncertainty modelling for differential gene expression estimation from RNA sequencing data | BMC Genomics | Full Text